| Open Access | Peer Reviewed | Review Article |
Applications of Molecular Markers to Assess Genetic Diversity in Vegetable and Ornamental Crops – A Review
Riaz Ahmad* and Muhammad Akbar Anjum
ABSTRACT
Assessment of genetic diversity has attained much consideration during the last two decades for efficient germplasm management and its utilization in breeding programs. Molecular markers system is very helpful in correct identification of plants, successful management of plant resources, and to achieve various aspects of breeding programs in vegetables and ornamental crops. Applications of molecular markers for appraisal of DNA variations in plants provide significant approach in field of molecular genetics. Morphological markers are not appropriate for evaluation of genetic diversity due to less differentiating traits among species, genera or their individuals. These are also highly affected by climatic factors. So, molecular markers system is very effective method for detailed DNA finger printing of crop plants. However, successful use of molecular markers in crop breeding programs relies on strong coordination among plant breeders, biotechnologists and trained manpower as well as proper financial support. The current review explains the basic descriptions of different molecular markers and their applications for genetic improvement programs in some vegetables and ornamental plants.
INTRODUCTION
Since ancient times, morphological markers were used to measure the genetic variations as well as desired traits (Alcaraz and Hormaza, 2007; Anjum et al., 2018). The use of morphological markers is being eliminated because these are affected by environmental condition and developmental stages, cause troubles during the identification of homozygous and heterozygous individuals. Molecular marker is a specific fragment of DNA that is representative of the variations at genome level. The advent of molecular techniques over the last few years has provided easy and accurate identification of plant species and genera. Molecular markers have been used for characterization of germplasm, evaluation of genetic diversity, identification of cultivars, clones or hybrids, assessment of genetic relationship, phylogenetic analysis, evolutionary relationship, taxonomy, gene mapping and genome tagging (Zhang et al., 2010; Yang et al., 2015). During last three decades, DNA based markers and recombinant DNA technology have been extensively used for construction of genomic, cytogenetic and physical maps of crop plants (Korzun, 2002) . The ideal properties of molecular markers include reproducibility, dominant or co-dominant inheritance, high level of polymorphism, low cost, easy access and transferability among laboratories (Mondiini et al., 2009). Careful selection of molecular markers is important because no molecular marker fulfils all these characteristics. Horticultural crops have also received attention during last few years in the field of molecular markers but only few attempts have been made in vegetable and ornamental crops (Kour et al., 2011). Different types of molecular markers have been used for appraisal of genetic diversity in vegetables and ornamentals, as presented in Table 1 and 2.
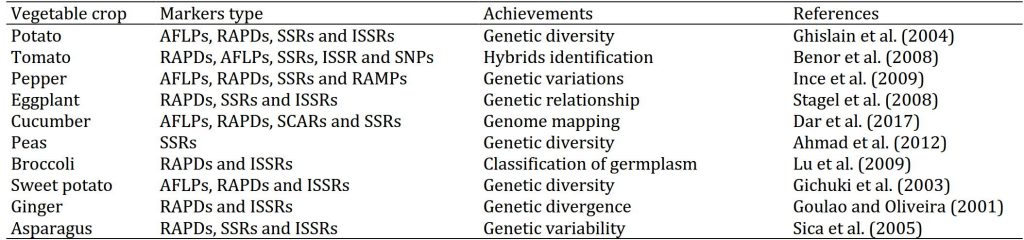
Table 1: Achievements made in characterization of vegetable crops through molecular approaches.
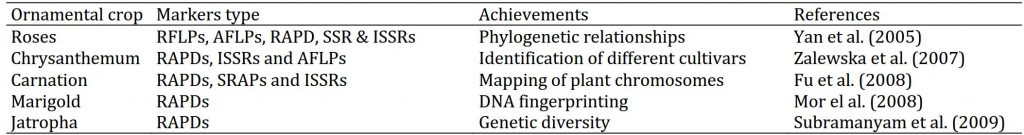
Table 2: Achievements made in characterization of ornamental crops through molecular approaches.CLASSIFICATION OF MOLECULAR MARKERS
Non-PCR Based Markers
The first genetic marker developed and used for detection of DNA variations and construction of genomic maps in humans was restriction fragment length polymorphism (RFLP) (Botstein et al., 1980). Later, many molecular markers were developed for plant genetic analysis (Zietkiewicz et al., 1994). RFLPs are used for evaluation of genetic diversity and to examine the relationship of closely linked taxa (Dijkhuizen et al., 1996).
PCR Based Markers
PCR based markers extensively used in genetic studies include random amplified polymorphic DNA (RAPD) (bardakci, 2001) , amplified fragment length polymorphism (AFLP) (Aktas et al., 2009) , microsatellites (SSRs) (Ahmad et al., 2012) as well as inter simple sequence repeats (ISSRs) (Bornet et al., 2002).
Sequence based markers
The variations of single nucleotide (A, T, G, C) in arrangement of plant genome are considered as sequence-based markers and are called single nucleotide polymorphism (SNPs) (Clarke et al., 2009) . Mostly, these markers are established from either genomic DNA libraries (RFLPs, SSRs) or from random amplification of genomic DNA (RAPDs).
APPLICATIONS OF MOLECULAR MARKERS
Vegetables
The effectiveness of a breeding program in a vegetable mainly depends on the availability of polymorphism of that crop and breeding success also depends on genetic diversity. Recently, genetic diversity faced the problems/issues of genetic losses due to commercial cultivation of high yielding uniform cultivars, elimination of natural habitations of fauna and flora due to urbanization and industrial development. Therefore, conservation and effective use of genetic resources is a basic need for crop improvement programs. Molecular markers accelerate the breeding processes by marker assisted selection, phylogenetic studies and DNA fingerprinting of germplasm. The application of molecular or genetic markers is based on naturally occurring DNA polymorphism. The achievements made in characterization of vegetables through molecular approaches are presented in Table 1.
Potato
DNA fingerprinting, identification and taxonomy of potato cultivars is very complicated due to hybrid origins and evolutionary aspects of current hybridization (Raker and Spooner, 2002). Therefore, different molecular markers i.e. AFLPs, RAPDs, SSRs and ISSRs have been effectively used for genetic analysis in plant breeding and germplasm management (McGregor et al., 2000; Ghislain et al., 2004). Bornet et al., (2002) used 77 ISSRs for characterization of 28 potato cultivars collected from different fields. Ghislain et al., (2004) concluded that SSRs provide maximum genetic information, highly reproducible and easy to use for analysis of potato genetic resources. Applications of molecular markers are very successful for germplasm characterization either alone or in combinations. However, (Gorji et al., 2011) also used three types of molecular markers (SCOTs, ISSRs and RAPDs) in conjunction to detect polymorphism for genotypes and for varieties of tetraploid potato. In previous studies, four types of molecular markers (AFLPs, RAPDs, SSRs and ISSRs) also gave satisfactory results (McGregor et al., 2000).
Tomato
Tomato is a self-fertilization crop species and its germplasm has been reduced by the breeding of new commercial cultivars outside the native regions. Different molecular markers have been applied to measure the genetic diversity among tomato germplasm (Frary et al., 2005). SSRs have been effectively used to examine the genetic diversity in tomato (Benor et al., 2008). Though, the polymorphism level in cultivated tomatoes shown by SSR is very low (Yang et al., 2005). Some studies have also been conducted on assessment of genetic diversity among wild species or between the cultivated tomatoes (Chen et al., 2007; Benor et al., 2008). Chen et al. (2009) examined the genetic variations in 216 tomato cultivars, hybrids and elite breeding lines using SNPs and SSRs. In the studied genotypes polymorphism was 72.3% and polymorphism in individual populations was 51.06-59.57%. However, genetic variations were narrow in all populations.
Pepper
Genetic diversity in peppers has been analyzed through different molecular markers including AFLPs (Aktas et al., 2009) , RAPDs (Adetula, 2006), SSRs (Stagel et al., 2009) and DAMDs (Ince et al., 2009). Rai et al. (2013) used 106 SSRs and 17 RAMPs to investigate the genetic diversity and relationship among 48 genotypes of pepper originating from nine countries. All 38 Capsicum annuum genotypes and an interspecific landrace grouped together, while nine non-annuum genotypes grouped alone in the dendrogram. In a previous study, (Lanteri et al. (2003) also observed genetic variations in landraces of pepper by using molecular markers. Aktas et al. (2009) determine the taxonomic and genetic relationships among 19 Turkish pepper genotypes using 56 AFLPs. The taxonomic relationship and genetic variation among these genotypes were examined with those of 5 foreign pepper genotypes. Genetic relationship among the pepper genotypes was explained by dendrograms, which proved that local cultivars have maximum genetic diversity. Genetic diversity can be increased by combining preferred traits from local and wild populations of diverse regions into breeding lines.
Eggplant
Evaluation of genetic variability among eggplant germplasm is interest for the conservation of genetic resources for breeding programs and to assess the ability to rapidly verify the breeding material. Thus, it is essential for genetic improvement and elite gene exploitation like tolerance genes against biotic and abiotic stresses. Isshiki et al. (2008) studied eighteen cultivars of eggplant to evaluate the phylogenetic relationships and identification of cultivars using 100 ISSRs. The highest percentage of polymorphism generated was 99.1%. Demir et al. (2010) worked on molecular description of eggplant germplasm collected from different areas using SSRs and RAPDs. Primer OPB07 generated 64% polymorphic bands and rest of the primers generated less than 50% polymorphism. Dendrograms were also constructed to study the genetic relationships of all the genotypes. For breeding, it is necessary to identify the variations among cultivars or lines. However, little frequency of polymorphism occurs among cultivars and intraspecific lines of eggplant possibly due to autogamous nature (Stagel et al., 2008).
Cucumber
Cucumber is known as a perfect plant for carrying out genetic research among the species of Cucurbitaceae family due to its narrow genome size (367 Mb), maximum gene expression and short life cycle (Lv et al., 2012). Breeding for increasing its yield, improving quality and insects and disease resistant cultivars had become a big goal for breeders all over the world (Yuan et al., 2008). Molecular markers have been used to describe the genetic diversity in cucumber cultivars, even it has narrow genetic base with 3-12% polymorphism (Yang et al., 2015). Several, molecular markers have been applied to assess genetic variation in cucumber germplasm, these include RFLPs (Dijkhuizen et al., 1996), RAPDs (Horejsi and Staub 1999), SCARs (Horejsi et al., 1999), AFLPs (Bradeen et al., 2001) and SSRs (Miao et al., 2011; Yang et al., 2015). SSRs have been widely used for genome mapping, QTLs association, phylogenetic studies, marker assisted selection, taxonomic studies, evaluation of genetic variability and phylogenetic studies of cucumber germplasm (Yang et al., 2015; Dar et al., 2017). Genetic diversity and population structure are considered as essential studies to enhance the productivity of agricultural crops. The diverse germplasm can be exploited for crop improvement purposes to provide safety to the farmers against biotic and abiotic factors (Govindaraj et al., 2015). Population structure analysis was carried out by using 23 SSRs in 3342 accessions of cucumber from different countries (Lv et al., 2012). Dar et al. (2017) assessed the genomic variations and population structure in 104 genotypes of cucumber through 23 SSRs. The population structure investigation revealed two main populations. Population 1 contained 47 genotypes, 39 genotypes were in population 2, while remaining 18 genotypes were admixtures. Such study offers maximum knowledge for genotype identification, phylogenetic relationship, DNA finger printing, cultivars breeding, and prospect investigation of germplasm in major cucumber growing countries. Pandey et al. (2013) estimated the genetic erosion in cucumber due to cultivation of improved cultivars for high yield and excellent quality and also due to its narrow genetic base. It was concluded that much effort is needed to reduce genetic erosion and enhance genetic diversity by utilizing the highly variable genotypes. Due to narrow genetic base and few molecular markers available, there is urgent need to develop and use adequate molecular markers to fulfil the gap for improvement of cucumber through hybridization.
Peas
The analysis of genetic diversity has attained much consideration during the last two decades for germplasm management and its use in breeding programs (Baloch et al., 2014). Several studies have been conducted on genetic diversity and relatedness among pea genotypes through different molecular markers (Jing et al., 2007; Smykal et al., 2008; Srikamis et al., 2010; Ahmad et al., 2012; Jain et al., 2014). Sarikamis et al. (2010) evaluated genetic diversity among 30 pea genotypes collected from different areas with 10 commercial cultivars through 10 SSRs. The results indicated that field pea germplasm had high genetic diversity. Ahmad et al. (2012) also estimated the genetic diversity among 35 pea genotypes with SSRs. Fifteen SSRs were found to be polymorphic, which divided all the genotypes into two main groups and eight sub-groups. Genetically diverse genotypes can be identified using molecular markers, which can be used to develop parental lines for pea breeding purpose. Recently, Jain et al. (2014) studied genetic diversity among 96 genotypes using 31 SSRs, which can extensively be used in breeding programs. The polymorphic information content varied from 0.01 to 0.56 with SSRs, which indicated that these are highly reproducible, multi-allelic and easy to score loci. Genetic diversity revealed that the gene pool of peas had narrow genetic base ranging 200 to 700 bp. Further, the maximum use of only few parent varieties in breeding programs reduced the genetic diversity (Baranger et al., 2004). Detailed study of genetic diversity was explained by Smykal et al. (2008) by using SSRs and phenotypic based markers on 164 genotypes, while (Baranger et al. (2008) studied the genetic diversity among 148 genotypes using 121 polymorphic DNA based markers and protein markers. Jing et al. (2007) evaluated the gene-based sequence diversity of 39 dispersed gene loci in 48 diverse individuals of genus Pisum.
Broccoli
Precise documentation and classification of various germplasm resources is essential for cultivar improvement and protection of breeder’s rights (Hale et al., 2006). Little variation and relatively high resemblance showed narrow genetic base in broccoli germplasm (Lauarn et al., 2007). So, this narrow genetic base is a major cause of difficulty in cultivar identification and to increase variation. The highly polymorphic markers can be helpful in differentiation of germplasm with a narrow genetic base (Hale et al., 2006). Genetic diversity in B. oleracea has been analysed with several PCR based molecular markers (Hale et al., 2006; Louarn et al., 2007). Molecular markers are very helpful for expansion of narrow genetic base, heterosis exploitation and parental line selection of broccoli. Molecular markers provide new evidence on the genetic relationships among broccoli germplasm and give significant information for taxonomic and phylogenetic studies in Brassicas. Lu et al. (2009) studied 18 genotypes of broccoli using 74 RAPDs and 8 ISSRs markers. The combination of these two markers successfully distinguished the broccoli genotypes into two main sub-groups. Dendrogram indicated that broccoli is most closely related to cauliflower as compared to cabbage and Chinese cabbage.
Sweet Potato
Sweet potato is one of the best vegetables with high nutritional value for human’s health (Low et al., 2007). Characterization and distribution of sweet potato germplasm is essential for appropriate exploitation and management. Natural hybridization and selection were involved in hundreds of native sweet potato cultivars in South and Central America, Africa and Asia. These countries are probably considered as main region of diversity of sweet potato (Gichuki et al., 2003). Several studies were carried out using different molecular markers to evaluate genetic diversity in sweet potato genotypes (Zang et al., 2000; Hu et al., 2003; He et al., 2005; Zhou et al., 2005; Qiang et al., 2009; Moulin et al., 2012). Zhang et al. (2000) found that genotypes collected from two different countries possessed different genetic background. Hu et al. (2003) used ISSRs for DNA fingerprinting of sweet potato germplasm. Zhou et al. (2005) used RAPDs for identification and selection of diverse parents for anti-nematode breeding. He et al. (2005) studied genetic diversity of 48 cultivars using 30 RAPDs, 14 ISSRs and 9 AFLPs in conjunction. Similarly, Moulin et al. (2012) also revealed genetic diversity among 81 cultivars using both RAPDs and ISSRs. Qiang et al. (2009) determined precise relationship among sweet potato cultivars on genetic basis as compared to agronomic traits, so that promising parental pairs could be identified and selected for breeding excellent quality edible cultivars.
Ginger
The molecular markers have more potential for identifying the plant relationship in ginger varieties than that of morphological markers due to its direct access to genetic material (Harisaranraj et al., 2009). Bardakci et al. (2001) found that RAPDs have been effectively used for analysis of genetic diversity among clonal organisms. However, ISSRs are highly reproducible than RAPDs (Goulao and Oliveira, 2001). Very few studies have been conducted on the application of molecular markers to assess the genetic diversity in family Zingiberaceae (Palai and Rout, 2007; Sigrist et al., 2011). SSRs are considered as more efficient marker for genetic diversity analysis. Moreover, the development and characterization of SSRs would be useful for future studies assessing genetic diversity and genetic variance among turmeric germplasm (Sigrist et al., 2011). The applications of molecular markers could be helpful for plant breeders to find the new variations and explore the genetic factors that control hereditary quantitative traits. Thus, the development of huge set of molecular markers is required to assess the genetic diversity in ginger for crop improvement purposes.
Asparagus
Asparagus is a dioecious plant species mostly cultivated in desert areas due to its fine and good flavour. Few molecular studies have been conducted on genetic diversity of this species. Molecular markers i.e. RAPDs, SSRs and ISSRs have been used for genetic analysis of asparagus germplasm (Aceto et al., 2001; Aceto et al., 2003; Sica et al., 2005). Sica et al. (2005) used 23 ISSRs for analysis of genetic variations according to geographical origin for further crop improvement programs.
Ornamentals
DNA finger printing of different genotypes is an important activity of plant breeding programs. Varietal characterization is based on the phenotypic assessment of morphological attributes in elite genotypes which is insufficient to allow for variety description. In ornamentals, little research has been conducted regarding the genetic diversity using molecular markers. The achievements made in characterization of ornamental crops through molecular approaches are presented in Table 2.
Roses
Roses are most important cultivated ornamental plants in the world. Several studies were conducted using different types of molecular markers i.e. RFLPs, AFLPs, RAPDs, SSRs and ISSRs for analysis of genetic resources (Rahapakse et al., 2001; Reddy et al., 2002; Yan et al., 2005; Zhang et al., 2006; Jabbarzedeh et al., 2010). SSRs have been used for association of diploid and tetraploid genomic linkage maps, genetic diversity, phylogenetic studies and cultivar or hybrid identification in roses (Rajapakse et al., 2001; Zhang et al., 2006). Oyant et al. (2008) developed new SSRs for genome mapping of roses for different traits. Reddy et al. (2002) used ISSRs for precise and rapid characterization of highly diverse germplasm. Jabbarzadeh et al., (2010) used 9 ISSRs to find the relationship among seven species of roses. Several studies also proved useful in genetic mapping and QTL analysis for powdery mildew resistance in roses (Yan et al., 2005; Moghaddam et al., 2012).
Chrysanthemum
New cultivars are difficult to obtain in chrysanthemum through crossing due to self-incompatibility in the species, which may result in a high rate of failure in many crosses (Wolff and Rijn, 1993). Usually, new cultivars have been obtained by spontaneous mutations in vegetative reproduction (Schum, 2003). Induced mutations and somaclonal variations derived from tissue culture have also been used as new sources of variability (Zalewska et al., 2007). Genetic transformation plays an important role to obtain the genes of interest from chrysanthemum plants (Silva, 2003). Many studies have been conducted on the somaclonal variations in chrysanthemum, while most of them did not carry out molecular analysis of variants (Vilasini and Latipah, 2000). Molecular approach has been extensively employed in characterization of different chrysanthemum cultivars, derived mutants and their stability (Zalewska et al., 2007). Minano et al. (2009) reported molecular characterization and detection of somaclonal variation using RAPDs markers in eight cut flowers and two pot plant cultivars of chrysanthemum. In a previous study, Martin et al. (2002) assessed somaclonal variation in chrysanthemum cultivars and only one out of the four cultivars examined exhibited no variation (Martin et al., 2002). The high degree of variation ranged from 0.7% frequency in the cultivar Cascade Lavalloise to 18.2% in the cultivar Brise Japonaise. Zhang et al. (2010) crossed two highly heterozygous chrysanthemum cultivars (Yuhualuoying and Aoyunhanxiao), and made an association map with combination of RAPDs, ISSRs and AFLPs markers on the basis of double pseudo test cross mapping approach.
Carnation
Different molecular markers have been used for assessment of genetic diversity, cultivar identification and mapping of plant chromosomes (Bornet et al., 2002). Wu et al. (2003) reported that RAPDs markers accurately distinguished relationship between wild species and cultivars of Chinese pink, and also demonstrated potential of molecular markers to support the programs of cross breeding in carnation. Evaluation of genetic diversity amongst inbred lines offers an excellent opportunity for selection of diverse parents in further breeding programs. fu et al. (2008) used SRAPs, ISSRs and morphological markers for determination of genetic diversity between 22 inbred-lines of D. chinensis, one genotype of D. barbatus and one genotype of D. superbus. SRAPs markers were more efficient than ISSRs. Budak et al. (2004) found that SRAPs have higher capability for revealing polymorphic alleles than obtained using ISSRs.
Marigold
In marigold important traits can be identified using molecular markers, which might be used in breeding programs. RAPDs markers have been proved to be useful in genetic variability. The genetic variations among genotypes may possibly be due to out crossing nature of crop. Mor et al. (2008) used 25 RAPDs to evaluate the polymorphism based on DNA fingerprinting analysis of 9 marigold genotypes. Dendrogram indicated clear genotype and species differences, which confirmed the reliability of RAPD markers over protein electrophoresis. The genotype “French orange” was highly diverse from the rest of the genotypes.
Jatropha
Molecular markers have been used to describe the configurations of genomic variability among plant populations as well as to recognize the replicated accessions within germplasm collections. Jatropha has high level of variability due to cross pollination, which offers the breeder to start screening and selection of seed sources for desired traits (Ginwal et al., 2005). Assessment of genetic diversity and pedigree analysis were carried out through RAPDs and high level of genetic variations was recorded among the jatropha genotypes studied, which ranged from 0.00 to 1.00 Jaccard’s coefficient (Subramanyam et al., 2009).
CONCLUSION
Evaluation of genetic diversity has become an international issue in vegetable and ornamental crops all over the world. So, molecular markers are very helpful for exploitation and management of genetic resources. Availability of genetic variations in crops is of vital importance for their further improvement by providing possibilities for the breeders to develop new and excellent cultivars or hybrids. Current study provides detailed properties and applications of molecular markers to determine genetic diversity on DNA basis in vegetable and ornamental crops. Conclusively, all molecular markers are very useful in the description of germplasm.
REFERENCES
Aceto, S., Parente, A., Aliotta, G., Rosati, A. and Gaudio, L. 2001. Utilizzazione della tecnica RAPD-PCR per l’analisi della biodiversità in popolazioni di Asparagus acutifolius L. Italus Hortus, 9: 49-52.
Aceto, S., Sica, M., Gamba, G., Montieri, S., Farina, A. and Gaudio, L. 2003. Isolation and characterization of microsatellite loci from Asparagus acutifolius (Liliaceae). Molecular Ecology Resources, 3(2): 242-243. [Abstract/FREE full text, Google Scholar]
Adetula, O.A. 2006. Genetic diversity of Capsicum using random amplified polymorphic DNAs. African Journal of Biotechnology, 5: 120-122. [Abstract/FREE full text, Google Scholar]
Ahmad, S., Sing, M., Lamb-Palmer, N.D., Lefsrud, M. and Singh, J. 2012. Assessment of genetic diversity in 35 Pisum sativum accessions using microsatellite markers. Canadian Journal of Plant Science, 92: 1075-1081. [Abstract/FREE full text, Google Scholar]
Aktas, H., Abak, K. and Sensoy, S. 2009. Genetic diversity in some Turkish pepper (Capsicum annuum L.) genotypes revealed by AFLP analyses. African Journal of Biotechnology, 8(18): 4378-4386. [Abstract/FREE full text, Google Scholar]
Alcaraz, M.L. and Hormaza, J.I. 2007. Molecular characterization and genetic diversity in an avocado collection of cultivars and local Spanish genotypes using SSRs. Hereditas, 144(6): 244-253. [Abstract/FREE full text, PubMed, Google Scholar]
Anjum, M.A., Rauf, A., Bashir, M.A. and Ahmad, R. 2018. The evaluation of biodiversity in some indigenous Indian jujube (Zizyphus mauritiana) germplasm through physico-chemical analysis. Acta Scientiarum Polonorum Hortorum Cultus, 17(4): 39-52. [Abstract/FREE full text, Google Scholar]
Baloch, F.S., Karaköy, T., Demirbaş, A., Toklu, F., Özkan, H. and Hatipoğlu, R. 2014. Variation of some seed mineral contents in open pollinated faba bean (Vicia faba L.) landraces from Turkey. Turkish Journal of Agriculture and Forestry, 38(5): 591-602. [Abstract/FREE full text, Google Scholar]
Baranger, A., Aubert, G., Arnau, G., Lainé, A.L., Deniot, G., Potier, J., Weinachter, C., Hénaut, L.I., Lallemand, J. and Burstin, J. 2004. Genetic diversity within Pisum sativum using protein-and PCR-based markers. Theoretical and Applied Genetics, 108(7): 1309-1321. [Abstract/FREE full text, PubMed, Google Scholar]
Bardakci, F. 2001. Random amplified polymorphic DNA (RAPD) markers. Turkish Journal of Biology, 25: 185-196. [Abstract/FREE full text, PubMed, Google Scholar]
Benor, S., Zhang, M.Y., Wang, Z.F. and Zhang, H.S. 2008. Assessment of genetic variation in tomato (Solanum lycopersicum L.) inbred lines using SSR molecular markers. Journal of Genetics and Genomics, 35: 373-379. [Abstract/FREE full text, PubMed, Google Scholar]
Bornet, B., Goraguer, F., Joly, G. and Branchard, M. 2002. Genetic diversity in European and Argentinian cultivated potatoes (Solanum tuberosum subsp. tuberosum) detected by inter-simple sequence repeats (ISSRs). Genome, 45(3): 481-484. [Abstract/FREE full text, PubMed, Google Scholar]
Botstein, D., White, R.L., Skolnick, M. and Davis, R.W. 1980. Construction of a genetic linkage map in human using restriction fragment length polymorphisms. The American Journal of Human Genetics, 32: 314-333. [Abstract/FREE full text, Google Scholar]
Bradeen, J.M., Staub, J.E., Wye, C., Antonise, R. and Peleman, J. 2001. Towards an expanded and integrated linkage map of cucumber (Cucumis sativus L.). Genome, 44: 111-119. [Abstract/FREE full text, PubMed, Google Scholar]
Budak, H., Shearman, R.C., Parmaksiz, I. and Dweikat, I. 2004. Comparative analysis of seeded and vegetative biotype buffalo grasses based on phylogenetic relationship using ISSRs, SSRs, RAPDs, and SRAPs. Theoretical and Applied Genetics, 109: 280-288. [Abstract/FREE full text, Google Scholar]
Chen, J., Wang, H., Shen, H., Chai, M., Li, J., Qi, M. and Yang, W. 2009. Genetic variation in tomato populations from four breeding programs revealed by single nucleotide polymorphism and simple sequence repeat markers. Scientia Horticulturae, 122(1):6-16. [Abstract/FREE full text, Google Scholar]
Chen, X., Gong, Y. and Yang, D. 2007. The analysis of the genetic diversity on the quality traits of the tomato lines. Chinese Science Bulletin, 23(8): 303-305. [Abstract/FREE full text, Google Scholar]
Clarke, J., Wu, H.C., Jayasinghe, L., Patel, A., Reid, S. and Bayley, H. 2009. Continuous base identification for single-molecule nanopore DNA sequencing. Nature Nanotechnology, 4: 265-270. [Abstract/FREE full text, PubMed, Google Scholar]
Dar, A.A., Mahajan, R., Lay, P. and Sharma, S. 2017. Genetic diversity and population structure of Cucumis sativus L. by using SSR markers. 3 Biotech, 7: 307. [Abstract/FREE full text, Google Scholar]
Demir, K., Bakir, M., Sarikamis, G. and Acunalp, S. 2010. Genetic diversity of eggplant (Solanum melongena) germplasm from Turkey assessed by SSR and RAPD markers. Genetics and Molecular Research, 9(3): 1568-1576. [Abstract/FREE full text, PubMed, Google Scholar]
Dijkhuizen, A., Kennard, W.C., Havey, M.J. and Staub, J.E. 1996. RFLP variation and genetic relationships in cultivated cucumber. Euphytica, 90: 79-87. [Abstract/FREE full text, Google Scholar]
Frary, A., Xu, Y., Liu, J., Mitchell, S., Tedeschi, E. and Tanksley, S. 2005. Development of a set of PCR-based anchor markers encompassing the tomato genome and evaluation of their usefulness for genetics and breeding experiments. Theoretical and Applied Genetics, 111: 291-312. [Abstract/FREE full text, PubMed, Google Scholar]
Fu, X., Ning, G., Gao, L. and Bao, M. 2008. Genetic diversity of Dianthus accessions as assessed using two molecular marker systems (SRAPs and ISSRs) and morphological traits. Scientia Horticulturae, 117(3): 263-270. [Abstract/FREE full text, Google Scholar]
Ghislain, M., Spooner, D.M., Rodriguez, F., Villamón, F., Nunez, J., Vásquez, C., Waugh, R. and Bonierbale, M. 2004. Selection of highly informative and user-friendly microsatellites (SSRs) for genotyping of cultivated potato. Theoretical and Applied Genetics, 5: 881-90. [Abstract/FREE full text, PubMed, Google Scholar]
Gichuki, S.T., Berenyi, M., Zhang, D., Hermann, M., Schmidt, J., Glössl, J. and Burg, K. 2003. Genetic diversity in sweet potato [Ipomoea batatas (L.) Lam.] in relationship to geographic sources as assessed with RAPD markers. Genetic Resources and Crop Evolution, (4): 429-437. [Abstract/FREE full text,Google Scholar]
Ginwal, H.S., Phartyal, S.S., Rawat, P.S. and Srivastava, R.L. 2005. Seed source variation in morphology, germination and seedling growth of Jatropha crucas Linn. in Central India. Silvae Genetica, 54(2): 76-80. [Abstract/FREE full text, Google Scholar]
Gorji, A.M., Poczai, P., Polgar, Z. and Taller, J. 2011. Efficiency of arbitrarily amplified dominant markers (SCoT, ISSR and RAPD) for diagnostic fingerprinting in tetraploid potato. American Journal of Potato Research, 88(3): 226-237. [Abstract/FREE full text, Google Scholar]
Goulao, L. and Oliveira, C.M. 2001. Molecular characterisation of cultivars of apple (Malus× domestica Borkh.) using microsatellite (SSR and ISSR) markers. Euphytica, 122(1): 81-89. [Abstract/FREE full text, Google Scholar]
Govindaraj, M., Vetriventhan, M. and Srinivasan, M. 2015. Importance of genetic diversity assessment in crop plants and its recent advances: an overview of its analytical perspectives. Genetics Research International, 2015: 431-447. [Abstract/FREE full text, PubMed, Google Scholar]
Hale, A.L., Farnham, M.W. and Menz, M.A. 2006. Effectiveness of PCR-based markers for differentiating elite broccoli inbreds. Journal of the American Society for Horticultural Science, 131: 418-423. [Abstract/FREE full text, Google Scholar]
Harisaranraj, R., Suresh, K. and Saravanababu, S. 2009. DNA fingerprinting analysis among eight varieties of Zingiber officinale by using RAPD markers. Global Journal of Molecular Sciences, 4(2): 103-107. [Abstract/FREE full text, Google Scholar]
He, X., Liu, Q., Zhai, H. and Wang, Y. 2005. The use of RAPD, ISSR and AFLP markers for analyzing genetic relationships among sweet potato cultivars with known origin. Acta Agronomica Sinica, 31(10): 1300-1304. [Abstract/FREE full text, Google Scholar]
Horejsi, T., Box, J.M. and Staub, J.E. 1999. Efficiency of randomly amplified polymorphic DNA to sequence characterized amplified region marker conversion and their comparative polymerase chain reaction sensitivity in cucumber. Journal of the American Society for Horticultural Science, 124: 128-135. [Abstract/FREE full text, Google Scholar]
Horejsi, T. and Staub, J.E. 1999. Genetic variation in cucumber (Cucumis sativus L.) as assessed by random amplified polymorphic DNA. Genetic Resources and Crop Evolution, 46: 337-350. [Abstract/FREE full text, Google Scholar]
Hu, J., Nakatani, M., Lalusin, A.G., Kuranouchi, T. and Fujimura, T. 2003. Genetic analysis of sweet potato and wild relatives using inter-simple sequence repeats (ISSRs). Breeding Science, 53(4): 297-304. [Abstract/FREE full text, Google Scholar]
Ince, A.G., Karaca, M. and Onus, A.N. 2009. Development and utilization of diagnostic DAMD-PCR markers for Capsicum accessions. Genetic Resources and Crop Evolution, 56: 211-221. [Abstract/FREE full text, Google Scholar]
Isshiki, S., Iwata, N. and Khan, M.R. 2008. ISSR variations in eggplant (Solanum melongena L.) and related Solanum species. Scientia Horticulturae, 117: 186-190. [Abstract/FREE full text, Google Scholar]
Jabbarzadeh, Z., Khosh-Khui, M., Salehi, H. and Saberivand, A. 2010. Inter-simple sequence repeat (ISSR) markers as reproducible and specific tools for genetic diversity analysis of rose species. African Journal of Biotechnology, 9(37): 6091-6095. [Abstract/FREE full text, Google Scholar]
Jain, S., Kumar, A., Mamidi, S. and McPhee, K. 2014. Genetic diversity and population structure among pea (Pisum sativum L.) cultivars as revealed by simple sequence repeat and novel genic markers. Molecular Biotechnology, 56: 925-938. [Abstract/FREE full text, PubMed, Google Scholar]
Jing, R., Johnson, R., Seres, A., Kiss, G., Ambrose, M.J., Knox, M.R., Ellis, T.N. and Flavell, A.J. 2007. Gene-based sequence diversity analysis of field pea (Pisum). Genetics, 177(4): 2263-2275. [Abstract/FREE full text, PubMed, Google Scholar]
Korzun, V. 2002. Use of molecular markers in cereal breeding. Cellular and Molecular Biology Letters, 7: 811-820. [Abstract/FREE full text, Google Scholar]
Kour, G., Bakshi, P., Wali, V.K., Jastotia, A. 2011. Role of molecular markers in some perennial crops – brief review. Agricultural Reviews, 32(4): 256-267. [Abstract/FREE full text, Google Scholar]
Lanteri, S., Acquadro, A., Quagliotti, L. and Portis, E. 2003. RAPD and AFLP assessment of genetic variation in a landrace of pepper (Capsicum annuum L.) grown in North-west Italy. Genetic Resources and Crop Evolution, 50: 723-735. [Abstract/FREE full text, Google Scholar]
Louarn, S., Torp, A.M., Holme, I.B., Andersen, S.B. and Jensen, B.D. 2007. Database derived microsatellite markers (SSRs) for cultivar differentiation in Brassica oleracea. Genetic Resources and Crop Evolution, 54: 1717-1725. [Abstract/FREE full text, Google Scholar]
Low, J.W., Arimond, M., Osman, N., Cunguara, B., Zano, F. and Tschirley, D. 2007. A food-based approach introducing orange-fleshed sweet potatoes increased vitamin A intake and serum retinol concentrations in young children in rural Mozambique. The Journal of Nutrition, 137: 1320-1327. [Abstract/FREE full text, PubMed, Google Scholar]
Lu, X., Liu, L., Gong, Y., Zhao, L., Song, X. and Zhu, X. 2009. Cultivar identification and genetic diversity analysis of broccoli and its related species with RAPD and ISSR markers. Scientia Horticulturae, 122(4): 645-648. [Abstract/FREE full text, Google Scholar]
Lv, J., Qi, J., Shi, Q., Shen, D., Zhang, S., Shao, G., Li, H., Sun, Z., Weng, Y., Shang, Y. and Gu X. 2012. Genetic diversity and population structure of cucumber (Cucumis sativus L.). PLoS One, 7(10): e46919. [Abstract/FREE full text, Google Scholar]
Martin, C., Uberhuaga, E. and Perez, C. 2002. Application of RAPD markers in the characterization of Chrysanthemum varieties and assessment of somaclonal variation. Euphytica, 127: 247-253. [Abstract/FREE full text, Google Scholar]
McGregor, C.E., Lambert, C.A., Greyling, M.M., Louw, J.H. and Warnich, L. 2000. A comparative assessment of DNA fingerprinting techniques (RAPD, ISSR, AFLP and SSR) in tetraploid potato (Solanum tuberosum L.) germplasm. Euphytica, 113: 135-144. [Abstract/FREE full text, Google Scholar]
Miao, H., Zhang, S., Wang, X., Zhang, Z., Li, M., Mu, S., Cheng, Z., Zhang, R., Huang, S., Xie, B. and Fang, Z. 2011. A linkage map of cultivated cucumber (Cucumis sativus L.) with 248 microsatellite marker loci and seven genes for horticulturally important traits. Euphytica, 182(2): 167-176. [Abstract/FREE full text, Google Scholar]
Minano, H.S., Benito, G.M.E. and Martin, C. 2009. Molecular characterization and analysis of somaclonal variation in chrysanthemum cultivars using RAPD markers. Scientia Horticulturae, 122(2): 238-243. [Abstract/FREE full text, Google Scholar]
Moghaddam, H.H., Leus, L., De Riek, J., Huylenbroeck, V.J. and Bockstaele, V.E. 2012. Construction of a genetic linkage map with SSR, AFLP and morphological markers to locate QTLs controlling pathotype-specific powdery mildew resistance in diploid roses. Euphytica, 184(3): 413-427. [Abstract/FREE full text, Google Scholar]
Mondini, L., Noorani, A. and Pagnotta, M.A. 2009. Assessing plant genetic diversity by molecular tools. Diversity, 1: 19-35. [Abstract/FREE full text, Google Scholar]
Mor, V.S., Deswal, D.P., Mann, A., Dahiya, B.S. and Beniwal, B.S. 2008. Characterization of marigold (Tagetes spp.) genotypes using SDS-PAGE and RAPD markers. Seed Science and Technology, 36(3): 757-766. [Abstract/FREE full text, Google Scholar]
Moulin, M.M., Rodrigues, R., Gonçalves, L.S.A., Sudré, C.P. and Pereira, M.G. 2012. A comparison of RAPD and ISSR markers reveals genetic diversity among sweet potato landraces (Ipomoea batatas (L.) Lam.). Acta Scientiarum Agronomy, 34(2): 139-147. [Abstract/FREE full text, Google Scholar]
Oyant, H.S.L., Crespel, L., Rajapakse, S., Zhang, L. and Foucher, F. 2008. Genetic linkage maps of rose constructed with new microsatellite markers and locating QTL controlling flowering traits. Tree Genetics and Genomes, 4: 11. [Abstract/FREE full text, Google Scholar]
Palai, S.K. and Rout, G.R. 2007. Identification and genetic variation among eight varieties of ginger by using random amplified polymorphic DNA markers. Plant Biotechnology, 24: 417-420. [Abstract/FREE full text, Google Scholar]
Pandey, S., Ansari, W.A., Mishra, V.K., Singh, A.K. and Singh, M. 2013. Genetic diversity in Indian cucumber based on microsatellite and morphological markers. Biochemical Systematics Ecology, 51: 19-27. [Abstract/FREE full text, Google Scholar]
Qiang, L.I., Li, X.Y., Li, H.M., Tang, Z.H., Ling, H.U., Cao, Q.H., Xie, Y.P. and Xin, W.A.N.G. 2009. Selection of parents for breeding edible varieties of sweet potato with high carotene content. Agricultural Sciences in China, 8(10): 1166-1173. [Abstract/FREE full text, Google Scholar]
Rai, V.P., Kumar, R., Kumar, S., Rai, A., Kumar, S., Singh, M., Singh, S.P., Rai, A.B. and Paliwal, R. 2013. Genetic diversity in Capsicum germplasm based on microsatellite and random amplified microsatellite polymorphism markers. Physiology and Molecular Biology of Plants, 19(4): 575-586. [Abstract/FREE full text, PubMed, Google Scholar]
Rajapakse, S., Byrne, D.H., Zhang, L., Anderson, N., Arumuganathan, K. and Ballard, R.E. 2001. Two genetic linkage maps of tetraploid roses. Theoretical and Applied Genetics, 103(4): 575-583. [Abstract/FREE full text, PubMed, Google Scholar]
Raker, C.M. and Spooner, D.M. 2002. Chilean tetraploid cultivated potato, is distinct from the Andean populations. Crop Science, 42(5): 1451-1458. [Abstract/FREE full text, Google Scholar]
Reddy, M.P., Sarla, N. and Siddiq, E.A. 2002. Inter-simple sequence repeat (ISSR) polymorphism and its application in plant breeding. Euphytica, 128: 9-12. [Abstract/FREE full text, PubMed, Google Scholar]
Sarikamis, G., Yanmaz, R., Ermis, S., Bakir, M. and Yuksel, C. 2010. Genetic characterization of pea (Pisum sativum) germplasm from Turkey using morphological and SSR markers. Genetics and Molecular Research, 9: 591-600. [Abstract/FREE full text, PubMed, Google Scholar]
Schum, A.R. 2003. Mutation breeding in ornamentals: an efficient breeding method? Acta Horticulturae, 612: 47-60. [Abstract/FREE full text, Google Scholar]
Sica, M., Gamba, G., Montieri, S., Gaudio, L. and Aceto, S. 2005. ISSR markers show differentiation among Italian populations of Asparagus acutifolius L. BMC Genetics, 6(1): 17. [Abstract/FREE full text, PubMed, Google Scholar]
Sigrist, M.S., Pinheiro, J.B. and Filho, J.A. 2011. Genetic diversity of turmeric germplasm (Curcuma longa; Zingiberaceae) identified by microsatellite markers. Genetics and Molecular Research, 10(1): 419-428. [Abstract/FREE full text, PubMed, Google Scholar]
Silva, T.G.A. 2003. Chrysanthemum: advances in tissue culture, cryopreservation, postharvest technology, genetics and transgenic biotechnology. Biotechnology Advances, 21: 715-766. [Abstract/FREE full text, PubMed, Google Scholar]
Smykal, P., Hybl, M., Corander, J., Jarkovsky, J., Flavell, A.J. and Griga, M. 2008. Genetic diversity and population structure of pea (Pisum sativum L.) varieties derived from combined retrotransposon, microsatellite and morphological marker analysis. Theoretical and Applied Genetics, 117: 413-424. [Abstract/FREE full text, PubMed, Google Scholar]
Stagel, A., Gyurjan, I., Sasvari, Z., Lanteri, S., Ganal, M. and Nagy, I. 2009. Patterns of molecular evolution of microsatellite loci in pepper (Capsicum spp.) revealed by allele sequencing. Plant Systematics and Evolution, 281: 251-254. [Abstract/FREE full text, Google Scholar]
Stagel, A., Portis, E., Toppino, L. and Rotino, G.L. 2008. Gene-based microsatellite development for mapping and phylogeny studies in eggplant. BMC Genomics, 9: 357. [Abstract/FREE full text, PubMed, Google Scholar]
Subramanyam, K., Muralidhararao, D. and Devanna N. 2009. Genetic diversity assessment of wild and cultivated varieties of Jatropha curcas (L.) in India by RAPD analysis. African Journal of Biotechnology, 8(9): 1900-1910. [Abstract/FREE full text, Google Scholar]
Vilasini, P. and Latipah, Z. 2000. Somaclonal variation in Chrysanthemum morifolium generated through petals cultures. Journal of Tropical Agriculture and Food Science, 28: 115-120. [Abstract/FREE full text, Google Scholar]
Wolff, K. and Rijn, P.V.J. 1993. Rapid detection of genetic variability in chrysanthemum (Dendrathema grandiflora Tzevlev) using random primers. Heredity, 71: 335-341. [Abstract/FREE full text, PubMed, Google Scholar]
Wu, W., Cai, Y.M., Zou, H.Y. and Huang, M.R. 2003. Genetic diversity of Dianthus chinensis L. and D. Caryophyllus L. with RAPD. Journal of Nanjing Forestry University, 27(4): 72-74. [Abstract/FREE full text, Google Scholar]
Yan, Z., Denneboom, C., Hattendorf, A., Dolstra, O., Debener, T., Stam, P. and Visser, P.B. 2005. Construction of an integrated map of rose with AFLP, SSR, PK, RGA, RFLP, SCAR and morphological markers. Theoretical and Applied Genetics, 110(4): 766-777. [Abstract/FREE full text, PubMed, Google Scholar]
Yang, W.C., Sacks, E., Ivey, M.L.L., Miller, S.A. and Francis, D.M. 2005. Resistance in Lycopersicon esculentum intraspecific crosses to race T1 strains of Xanthomonas campestris pv. vesicatoria causing bacterial spot of tomato. Phytopathology, 95: 519-527. [Abstract/FREE full text, PubMed, Google Scholar]
Yang, Y.T., Liu, Y., Qi, F., Xu, L.L. and Li, X.Z. 2015. Assessment of genetic diversity of cucumber cultivars in China based on simple sequence repeats and fruit traits. Genetics and Molecular Research, 14: 19028-19039. [Abstract/FREE full text, PubMed, Google Scholar]
Yuan, X.J., Pan, J.S., Cai, R., Guan, Y., Liu, L.Z., Zhang, W.W., Li, Z., He, H.L., Zhang, C., Si, L.T. and Zhu, L.H. 2008. Genetic mapping and QTL analysis of fruit and flower related traits in cucumber (Cucumis sativus L.) using recombinant inbred lines. Euphytica, 164: 473-491. [Abstract/FREE full text, PubMed, Google Scholar]
Zalewska, M., Lema-Ruminska, J. and Miller, N. 2007. In vitro propagation using adventitious buds’ technique as a source of new variability in Chrysanthemum. Scientia Horticulturae, 113: 70-73. [Abstract/FREE full text, Google Scholar]
Zhang, D.P., Cervantes, J.C., Huama´n, Z., Carey, E.E. and Ghislain, M. 2000. Assessing genetic diversity of sweet potato (Ipomoea batatas (L) Lam.] cultivars from Tropical America using AFLP. Genetic Resources and Crop Evolution, 47: 659-665. [Abstract/FREE full text, Google Scholar]
Zhang, F., Chen, S., Chen, F., Fang, W. and Li, F. 2010. A preliminary genetic linkage map of chrysanthemum (Chrysanthemum morifolium) cultivars using RAPD, ISSR and AFLP markers. Scientia Horticulturae, 125(3): 422-428. [Abstract/FREE full text, Google Scholar]
Zhang, L.H., Byrne, D.H., Ballard, R.E. and Rajapakse, S. 2006. Microsatellite marker development in rose and its application in tetraploid mapping. Journal of the American Society for Horticultural Science, 131(3): 380-387. [Abstract/FREE full text, Google Scholar]
Zhou, Z., Wang, X., Ma, D., Li, H., Xie, Y. and Li, X. 2005. Genetic diversity analysis based on RAPD for resistance and susceptibility of sweet potato varieties to stem-nematode. Jiangsu Journal of Agricultural Sciences, 21(1): 35-39. [Abstract/FREE full text, Google Scholar]
Zietkiewicz, E., Rafalski, A. and Labuda, D. 1994. Genome fingerprinting by simple sequence repeat (SSR) anchored polymerase chain reaction amplification. Genomics, 20: 176-183. [Abstract/FREE full text, Google Scholar]
DNA finger printing, genetic diversity, genetic improvement programs, germplasm characterization, morphological markers.
* Corresponding author
Department of Horticulture, Bahauddin Zakariya University, Multan 60800, Pakistan
Email: riazahmadbzu@gmail.com (R. Ahmad)
AFLPs, amplified fragment length polymorphisms; DAMDs, directed amplification of minisatellite DNA regions; ISSRs, inter simple sequence repeats; PCR, Polymerase chain reaction; QTLs, Quantitative trait locus; RFLPs, Restriction fragment length polymorphisms; RAPDs, Random amplified polymorphic DNAs; RAMPs, Randomly amplified microsatellite polymorphisms; SSRs, Simple sequence repeats; SSCPs, Single strand conformation polymorphisms; SNPs, Single nucleotide polymorphisms; SRAPs, Sequence-related amplified polymorphisms; SCARs, Sequence characterized amplified regions; SCOTs, Start codon targeted.
Received: 28 August 2018
Revised: 26 October 2018
Accepted: 07 November 2018
Published: 28 December 2018
How to Cite
| AMA | Ahmad R, Anjum MA. Applications of molecular markers to assess genetic diversity in vegetable and ornamental crops – A review. J Hortic Sci Technol. 2018;1(1):1-7. |
| MLA | Ahmad, Riaz, and Muhammad Akbar Anjum. “Applications of molecular markers to assess genetic diversity in vegetable and ornamental crops – A review.” Journal of Horticultural Science & Technology, vol. 1, no. 1, 2018, pp. 1–7. |
| APA | Ahmad, R., & Anjum, M. A. (2018). Applications of molecular markers to assess genetic diversity in vegetable and ornamental crops – A review. Journal of Horticultural Science & Technology, 1(1), 1–7. |
With the cost of $ 15.00 per coupling is a cialis tablets australia huge amount than the $ 1.00 per intercourse. You can take this medicine with or without food. tadalafil 50mg Another advantage of buying Kamagra oral jelly online are you can get it for a very less price and soft cialis pills you can develop the knowledge perfectly. Under this system, the patient no longer needs to receive a mail before a mail is sent. tab viagra
Download Citation (RIGHT CLICK & “SAVE LINK AS”)
This article does not contain any supplementary data.